Bioinformatics software plays a vital role in transforming raw biomedical data into valuable biological insights.
With the explosion of biomedical data—thanks to advancements like whole-genome sequencing—the need for powerful bioinformatics tools is more important than ever.
But building this type of software isn’t a walk in the park.
It requires expertise in biology, computer science, and statistics. Plus, biological data comes in all sorts of formats, making it tricky to analyze.
And let’s not forget—the software needs to be user-friendly, especially for biologists who aren’t programmers.
Despite these challenges, the payoff is huge.
A well-designed bioinformatics tool can drive groundbreaking discoveries in healthcare, agriculture, and beyond.
This guide will walk you through everything you need to know to build successful bioinformatics software.
Over 90% of the world’s data has been generated in the last two years, much from life sciences. Bioinformatics is key to decoding this data. If you’re passionate about tech and biology, now is the time to develop impactful bioinformatics software!
Bioinformatics Software: The Backbone of Modern Biological Research
Bioinformatics software helps scientists make sense of biological data.
It’s used to analyze big datasets like DNA sequences, protein structures, and gene patterns.
In simple terms, it connects biology with technology, turning complex data into clear insights.
With this software, scientists can run complex calculations, visualize data, and even predict how genes or proteins will behave.
It’s widely used in areas like genomics, drug discovery, and personalized medicine.
In short, bioinformatics software makes it easier to understand the biological world.
It helps researchers unlock insights that could lead to important breakthroughs in science and medicine.
Market Stats & Growing Role of AI and Machine Learning in Bioinformatics
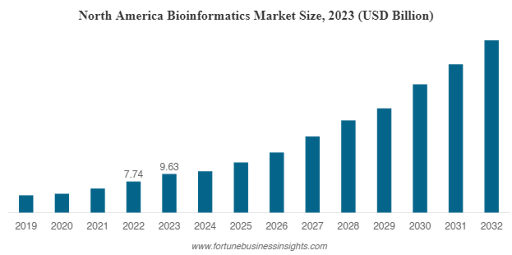
The bioinformatics market is booming.
In 2023, it was valued at $20.72 billion, and by 2032, it’s expected to reach $94.76 billion, growing at a rate of 17.6% per year.
One key driver of this growth is the use of artificial intelligence (AI) and machine learning (ML).
These technologies help researchers find patterns in large, complex datasets.
This speeds up drug discovery, improves prediction models, and supports personalized medicine.
For example, in July 2023, Genesis Healthcare in Japan launched GenesisGaia, a platform using AI for genomic data.
Similarly, FOXO Technologies introduced AI-powered bioinformatics services to push breakthroughs in biotechnology and epigenetic research.
AI and ML are transforming bioinformatics, and their impact on drug discovery and personalized healthcare is huge.
Best Practices for Bioinformatics Software Development
Bioinformatics software is designed to address a wide range of biological research questions. Here are some of the most common tasks that bioinformatics tools can help with:
1. Sequence Analysis
- Sequence alignment: Comparing sequences to identify similarities and differences.
- Motif finding: Locating specific patterns or motifs within sequences.
- Homology searching: Identifying related sequences in databases.
- Multiple sequence alignment: Aligning multiple sequences to study conserved regions.
2. Genome Assembly
- Short read assembly: Reconstructing genomes from short DNA sequences.
- De novo assembly: Assembling genomes without a reference genome.
- Genome annotation: Identifying genes, regulatory regions, and other genomic features.
3. Gene Prediction
- Ab initio prediction: Predicting genes based on computational models.
- Comparative gene prediction: Using conserved regions to identify genes.
- Gene expression analysis: Quantifying gene activity.
4. Phylogenetic Analysis
- Tree building: Constructing evolutionary trees to infer relationships between organisms.
- Phylogeny visualization: Representing evolutionary relationships graphically.
- Bootstrapping and other statistical methods: Assessing the reliability of phylogenetic trees.
5. Protein Structure Prediction
- Homology modeling: Predicting protein structures based on similar proteins.
- Ab initio prediction: Predicting protein structures from amino acid sequence alone.
- Molecular dynamics simulations: Studying protein dynamics and interactions.
6. Drug Discovery
- Virtual screening: Identifying potential drug candidates from large databases.
- Molecular docking: Predicting the binding affinity of molecules to target proteins.
- Pharmacophore modeling: Identifying common structural features of bioactive molecules.
7 Key Features to Include in Bioinformatics Software
When building bioinformatics software, there are several important features to consider. These will help ensure that the software is efficient, flexible, and secure.
1. Scalability
Make sure your software can handle large datasets. It should process data quickly without slowing down. This allows researchers to explore complex biological questions without running into performance issues.
2. Modularity
Design your software in a modular way. This means users can add or remove features as needed. It makes the software more flexible and easy to adapt as research needs evolve.
3. Interoperability
Your software should work well with other bioinformatics tools and databases. By supporting common data formats, users can easily exchange information across platforms.
4. Efficiency
Focus on creating efficient algorithms. This speeds up data processing and reduces the strain on resources, helping researchers get results faster.
5. Data Security
Protect sensitive biological data by including strong security features. Encryption, access controls, and regular audits are key to maintaining trust and ensuring privacy.
6. Workflow Automation
Automate repetitive tasks like data processing and analysis. This saves time and reduces errors, allowing researchers to focus on interpreting results.
7. Collaboration Tools
Include version control and collaboration features. These tools make it easy for teams to work together, track changes, and ensure that research is reproducible.
The Software Development Life Cycle in Bioinformatics
Developing bioinformatics software follows a structured process called the Software Development Life Cycle (SDLC). This cycle helps create reliable software that meets users’ needs. Here’s a simple breakdown of the key stages:
Choosing the Right Tech Stack for Bioinformatics Software
Selecting the right technology stack is vital for the success of your bioinformatics software.
It impacts scalability, performance, and flexibility. Here’s a quick overview of essential categories and options to consider:
- Programming Languages: Python, R, Java, C/C++, Perl
- Web Frameworks: Django, Flask, Shiny, Spring Boot, Ruby on Rails
- Data Storage: MySQL, PostgreSQL, MongoDB, SQLite, Oracle
- Bioinformatics Tools: BioPython, Bioconductor (for R), EMBOSS
- Cloud Platforms: Amazon Web Services (AWS), Google Cloud Platform (GCP), Azure
- Version Control: Git, SVN
- Data Visualization: Matplotlib, Seaborn, ggplot2, Plotly, D3.js
- Containerization Technology: Docker, Singularity
Each category offers various options, and the right choice depends on your project needs and team expertise.
Case Studies of Successful Bioinformatics Software Projects
Let’s take a look at two successful bioinformatics software projects that have made a significant impact in the field: clusterProfiler and CIRCexplorer2. These tools address specific challenges and help advance biological research.



